There's a category of experiment that R&D biologists still have a hard time automating. Why?
Definition: by “robot” we mean the entire functionality of a liquid handler or dispenser, specifically its ability to move and manipulate liquids.
Life science R&D lives and dies on the success of its experimentation, and this experimentation often relies on automated liquid handling robots. But an entire category of experiment isn’t that easy to automate. Even if they do get automated, they typically miss out on key benefits of automation, such as reproducibility, transferability, and productivity gains. It’s like this because of how most automation processes work today—they focus on moving the robot.
While this approach works in many other scenarios, it fails when experiments that have at least 2 of these 4 characteristics:
- Variable: the experimental conditions or the overall protocol are frequently changed
- Multifactorial: they study the intricacies of interactions between different factors
- Small-scale: they are run in the early stages of an experimental campaign before protocols are scaled up
- Emergent: protocols may need to be adapted depending on the results
In our experience, these experiments are especially prevalent in process development for modern therapeutic modalities such as biologics, where processes are less established. They are also common in assay development, where a protocol must be designed and then optimized over a multitude of experimental factors.
Despite often being small-scale, these experiments can be highly impactful to the overall outcomes of a drug discovery campaign because they are run in its early stages. More effective optimisation of an assay for High Throughput Screening (HTS) can result in better candidate selection or cost savings on expensive reagents. That’s because a better Z’ can lead to fewer false positives and negatives, or conditions can be found which yield a good enough Z’ while using less material. Iterating on a protocol during process development faster and with greater flexibility can result in substantially faster time to market, or better yield of a pharmaceutical.
Why are experiments like this so hard to automate? What would make it easier? In this article, we’ll look at the following:
- Liquid handling automation’s 4 main problems, and why they aren’t solved yet
- The limitations of robot-oriented lab automation
- What sample-oriented lab automation is, how it works, and what it makes possible
Let’s get into it.
Liquid handling automation: a 4-part problem
We can look at liquid handling automation as 4 steps, each with their own problem that needs solving:
- The execution of experimental protocols… [Problem 1: how do we represent protocols in a way that is unambiguous, executable by automation equipment, and readily understood by our collaborators?]
- …with speed, high throughput, and/or high accuracy… [Problem 2: how do we optimize these protocols for execution with speed, accuracy, and reproducibility against available equipment?]
- …to manipulate and analyze samples… [Problem 3: how do we track and understand the manipulation, transformation, and movement of the samples within these automated protocols?]
- …with the aim of gathering rich experimental data. [Problem 4: how do we gather data from and about our experiments, including analytical measurements and the corresponding experimental factors? How do we use it to build rich datasets that adhere to FAIR data principles without significant manual intervention?]
Each of these problems is harder to solve the more complex and unique an experiment is. If an experiment rarely changes, isn’t too complicated, runs at scale, and uses only 1 or 2 factors, most of the effort goes towards optimizing protocol execution and putting together suitable data analysis tools. We write our automation scripts and build these tools once, and are then able to repeat the experiment as frequently as required.
But if my experiment changes all the time, develops unpredictably as it progresses, runs only at a small scale, all while studying dozens of factors at many different levels… the effort required to automate may outweigh the perceived benefits. In such scenarios, scientists typically revert to manual execution and less effective experimental methods altogether, such as optimizing one factor at a time (OFAT).
The limits of robot-oriented lab automation (ROLA)
The traditional approach to these problems requires meticulous translation of a protocol into detailed robot instructions, specifying the source and destination of each liquid transfer along the way. Since this approach focuses heavily on the robot’s actions, we refer to this as robot-oriented lab automation (ROLA). It often requires a team of lab scientists, automation engineers, and data scientists to build a shared mental model and a blueprint of the protocol. This blueprint then gets translated into automation scripts, worklist-generating programs, and spreadsheets comprising a tailored toolkit.
These tools ideally offer flexibility for various experimental conditions: pH, reagent concentrations, incubation times, and more. But here's the catch: the more flexibility you need, the harder it is to create this toolkit. Even after setting the robots in motion, aligning experimental readings with the specific conditions for each sample is a whole other challenge.
All in all, this process comes with 3 significant downsides:
- Low economic feasibility: pouring so much effort into automation often doesn't add up, especially for highly variable or multi-factorial experimental protocols. Unless the protocol will be repeated many, many times, the cost of creating the tools can be prohibitive.
- Information management nightmare: artifacts from this process scatter across multiple software tools. This makes storage, backup, and change-tracking a colossal task.
- Difficult collaboration: each toolkit is custom-built for a specific protocol, meaning knowledge doesn't easily transfer from one set of tools to another. Moreover, understanding the workings of a toolkit and the data it generates is not straightforward without assistance from its creators or proficient users.
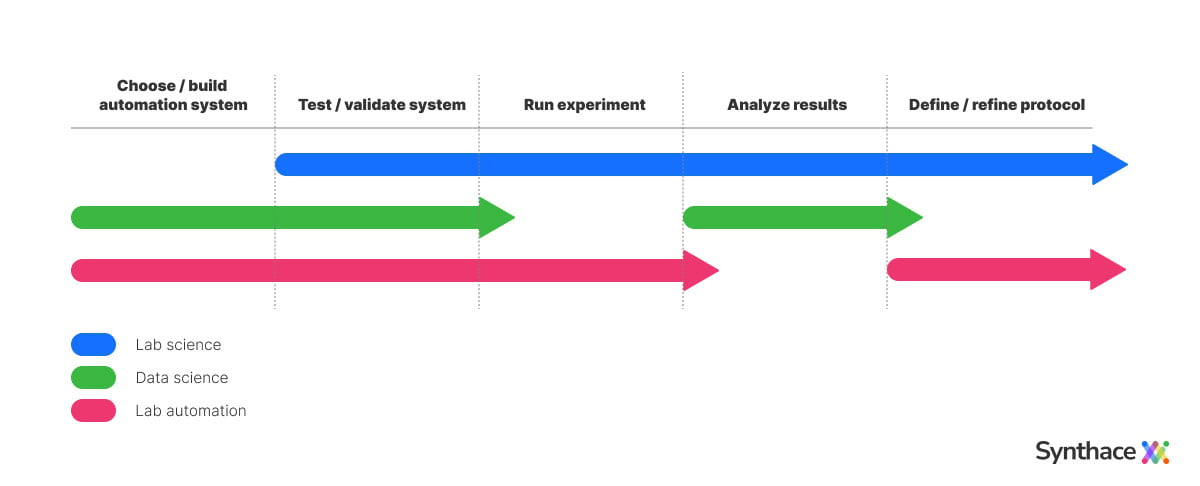
Larger projects can span multiple months and require collaboration from different teams. Often there's a disconnect between the lab scientists who design the process based on scientific considerations, lab automation engineers who are more concerned with the challenges of its physical implementation, and data scientists who are focused on building the pipeline to analyze the acquired data.
Communication is often reliant on a mixture of vague whiteboard sketches and laborious handoff documentation. This leads to a prolonged back-and-forth at each stage of the project. Even after a system is up and running, subsequent refinements to it are equally challenging to deliver.
Bottom line: for a certain class of experiments, ROLA won't be your best bet.
A different approach: sample-oriented lab automation (SOLA)
Sample-oriented lab automation (SOLA) works at a higher level of abstraction. With abstraction, we spend more time dealing with an idea rather than a thing. It works at relative levels:
- “I want to run this protocol” is more abstract than…
- “I want to combine these samples” is more abstract than…
- “I want to aspirate from A1, then dispense to B1.”
When we try to automate those variable, emergent, multifactorial, and small-scale experiments with low-level ROLA abstraction, we spend more time working on the thing than the idea. Writing low-level scripts from scratch each time is impractical for these experiments. It’s also pretty unpleasant.
Rather than micromanaging different pipetting robots, we could instead tell a software package what we want to happen to our samples. This software could then convert those instructions into low-level code for different robots, then structure all the resulting data back against the experimental conditions for each sample.
A SOLA approach allows a scientist to automate experiments by first defining sets of samples and then performing logical operations on them using familiar terminology. They can build their protocol in terms of configurable steps (we call them “elements”) within a visual programming environment, allowing them to focus on what’s happening to their samples instead of how their robots are moving.
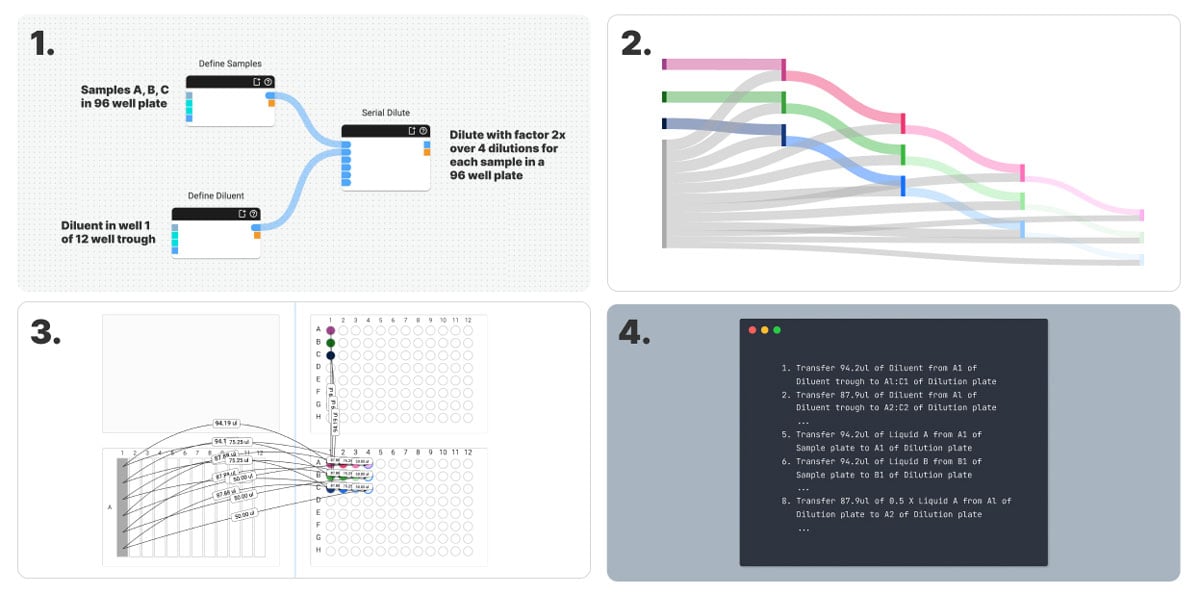
Depicted above are four different representations of a serial dilution protocol, from most to least abstract. Firstly we have (1) the SOLA workflow with 3 elements: 2 to define the samples and diluent and one to configure the serial dilution. Next we have (2) representations of the sample flow and (3) the transfers required to achieve this sample flow over a set of plates on the deck of our liquid handler. Finally, we have (4) a list of robot instructions down to individual aspirate and dispense operations. It’s evident that as we move down the ladder of abstraction, the scientific intent of our protocol becomes more difficult to infer from the representation.
By using a software package to perform the translation from the sample-oriented workflow down to robot instructions, we can more clearly communicate our intentions to our collaborators. This approach also alleviates the cognitive load of the traditional robot-oriented approach and builds the foundation for a structured, computable model of biological experiments themselves. The experimental conditions for each sample can also be automatically propagated throughout, along with any metadata about them, meaning we can automatically align these with analytical measurements.
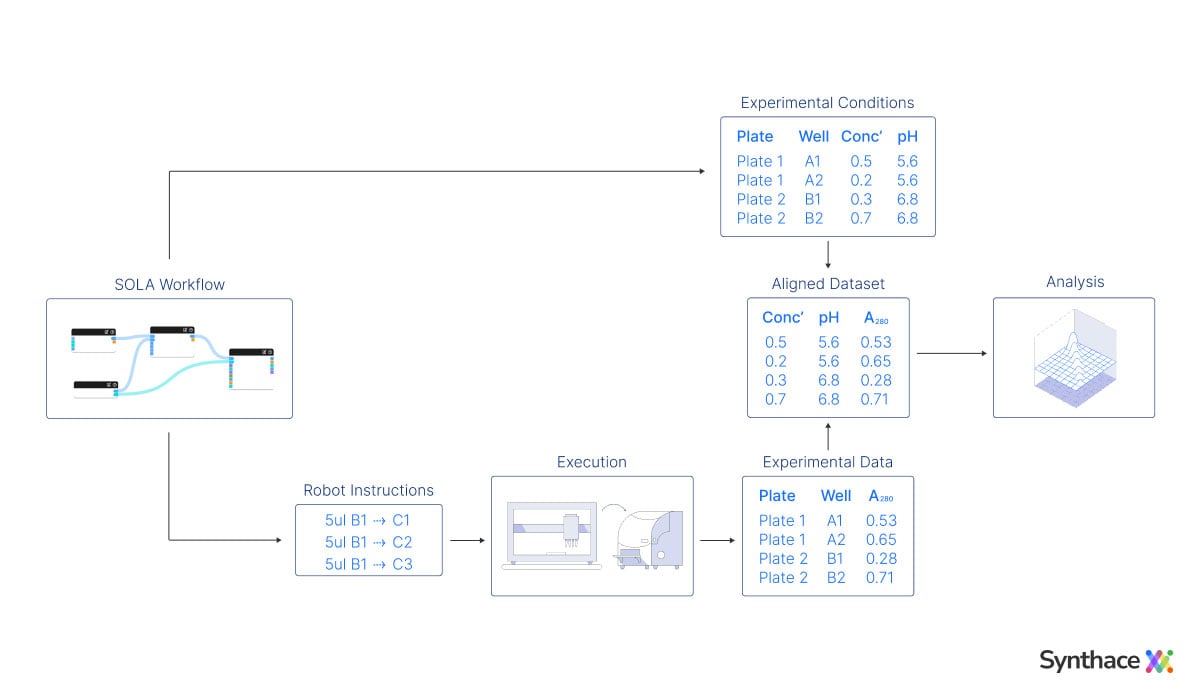
Here are those 4 liquid handling automation problems again, and how SOLA solves them on the Synthace platform:
- Problem 1—protocol creation: we describe a protocol by defining liquids and manipulations done to them to construct a workflow
- Problem 2—protocol automation: the platform converts the workflow into instructions for liquid handling equipment
- Problem 3—sample provenance: the samples are defined in the context of the automation instructions, and so get tracked through the protocol as it runs
- Problem 4—data and metadata: the experimental design, sample definitions, automation instructions, experimental data, and metadata are aligned in one place
What a SOLA approach makes possible
SOLA workflows give a precise representation of biological experiments, emphasizing sample flow and leaving no room for ambiguity. This makes them much more legible than their ROLA counterparts, enabling more effective collaboration between scientists, lab automation experts, and data scientists.
What's more, SOLA isn't shackled to any specific robot. It's adaptable, operating at a higher level of abstraction allowing straightforward method transfer from one robot to another. SOLA on Synthace isn’t limited to lab automation either. It can also generate step-by-step instructions for manual execution, making it possible for labs to start their journey toward automation before even having access to a liquid handling robot.
Here are 3 of the main impacts of SOLA when adopted:
- Makes it feasible to automate tasks that would otherwise be done manually. Thus experimental protocols are unambiguously documented, leading to improved reproducibility and datasets with rich metadata. Many Synthace users start doing more routine tasks on their liquid handlers instead of with manual pipetting.
- It streamlines the process of automating protocols by providing the ability to iterate on them with greater ease and flexibility. We see this among Synthace users who specialize in miniaturized purification (i.e. using RoboColumns or filter plates on Tecan liquid handlers). They are often able to optimize a protocol more quickly, leading to faster time to market, or optimize their protocol further, leading to better yield of a precious pharmaceutical.
- Most excitingly, it unlocks more effective experimental methods altogether, such as complex multifactorial experiments. We often find this among scientists using Synthace for assay development, where these improved methods can be truly transformative.
Shifting to a sample-oriented approach in lab automation can be a difficult step at first. It requires some learning, some unlearning, and some trial and error in the early days. For many, there’s an “aha!” moment during the early stages.
There is a possibility, at some point in the far future, that an even higher level of abstraction may eventually be possible. But, right now, the more teams we can support in their move toward a sample-oriented view of the world, the better.
Anton Tcholakov
Anton Tcholakov is a Senior Software Engineer, helping accelerate life science R&D at Synthace. He has 7 years' experience in making advanced R&D technologies easier to use and adopt, across the life sciences, electronics, and automotive industries.



