We recently combined OpenAI’s ChatGPT with the Synthace platform to design protocols for biological experiments and automate lab work. In this blog, we’ll get into the details of how we were able to do this, explore some possibilities and limitations, and indulge in some (brief) speculation about how this might develop in the near future. Toward the end, we also answer some FAQs and give details about how you can collaborate with us.
A quick disclaimer for those who don’t read further: this is a working proof of concept, but isn’t widely available on our platform right now.
Before we get into it, here’s a quick summary:
- We’ve connected OpenAI’s ChatGPT API with our digital experiment platform
- We’ve used this interface to define and run biology experiments on different automation equipment in our lab
- It works by connecting OpenAI’s API (to query GPT-3.5) and the internal Synthace API
- This is currently a proof-of-concept and is not yet commercially available, but we’ve already had strong interest from those to whom we’ve given previews
Here’s a short video showing how it works:
It makes what would otherwise be an incredibly difficult, time-consuming process look easy. But there’s a lot going on underneath the hood, all of which needs to work before the AI can act at this level of abstraction.
The promise of Large Language Models (LLMs) and lab automation
A couple of years ago, our UX designer organized a design sprint where we brainstormed ideas for how to build the user interface of our digital experiment platform. My favorite idea was a simple chat-based interaction that would guide you through the experiment setup.
It seemed like a long way off at the time. Just a short while later, it seems to be here.
A note of caution, though: natural language, on its own, isn’t enough to unambiguously represent biological protocols. But then again, pure code can be hard to rapidly understand. Humans can easily understand the former, albeit with a lot of ambiguity. On the other hand, machines can act powerfully with unambiguous code but it’s harder for humans to work with than with natural language.
For a very long time, the most common workaround to this problem has been graphical user interfaces (GUIs) bolted onto lab robots. Erika Alden DeBenedictis puts it well in her blog:
"The need to write code has always been a huge barrier that prevents lab automation from becoming widespread. Historically, biologists weren’t taught programming. As a result, robot manufacturers in life science usually present users with GUI interfaces. Short-term, GUIs are a win that allow users to quickly get robots to do simple tasks in a no-code setting. Long term, GUIs shoot lab automation in the foot. Protocols developed in a GUI just can’t be that sophisticated or scalable. This raises a big question: if lab automation is ever going to change how we do biology research, how do we resolve the tension between the GUI vs coding interface?"
This tension leaves biologists in one of four scenarios:
- Don’t automate; use natural language and manual experiments
- Automate, with limitations, using GUI interfaces on robots
- Learn to code the robots yourself
- Work in tandem with an automation scientist/engineer
The more advanced you get with code, the more automation power you can use… at the risk of spending more time working on your code than your science. But, as DeBenedictis continues, “Fully-automated biology faces the programming-barrier-to-entry on steroids. A lot of first users [at an automation challenge] were learning programming, laboratory robotics, and science all at the same time. The learning curve was steep to the point of being impossible to surmount.”
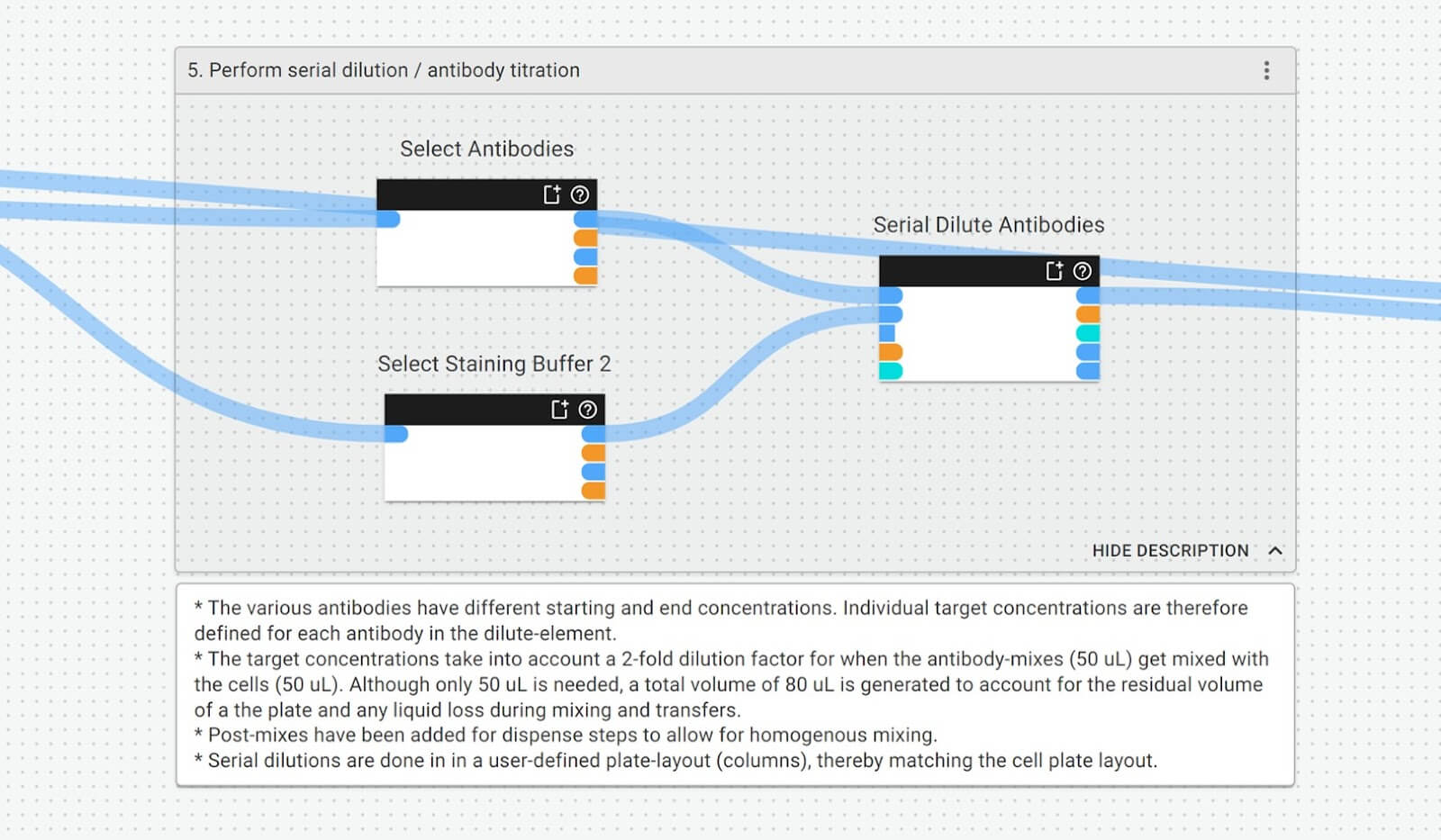
A typical view showing part of a protocol on the Synthace platform
Combining natural language and a GUI should, in theory, move towards resolving the problem if done at a high enough level of abstraction. Before ChatGPT was even announced, this is what our platform did and still does with our workflow builder: a visual and natural language representation of protocols, acting as an abstraction of the code underneath it, against which we can run automation equipment, then collect detailed lab data and metadata. This forms part of what we call a digital experiment model.
Adding ChatGPT to this model, however, seems to create something entirely new altogether. Let’s look at it in more detail.
How we use ChatGPT to design and run biology experiments
This prototype integration is based on communication between two services:
- OpenAI’s API to query the GPT-3.5 model used by ChatGPT (unless we’re specifically referring to the model, we'll subsequently refer to this as ChatGPT)
- Our internal Synthace API that allows the creation of workflows and simulations against the Synthace planner programmatically
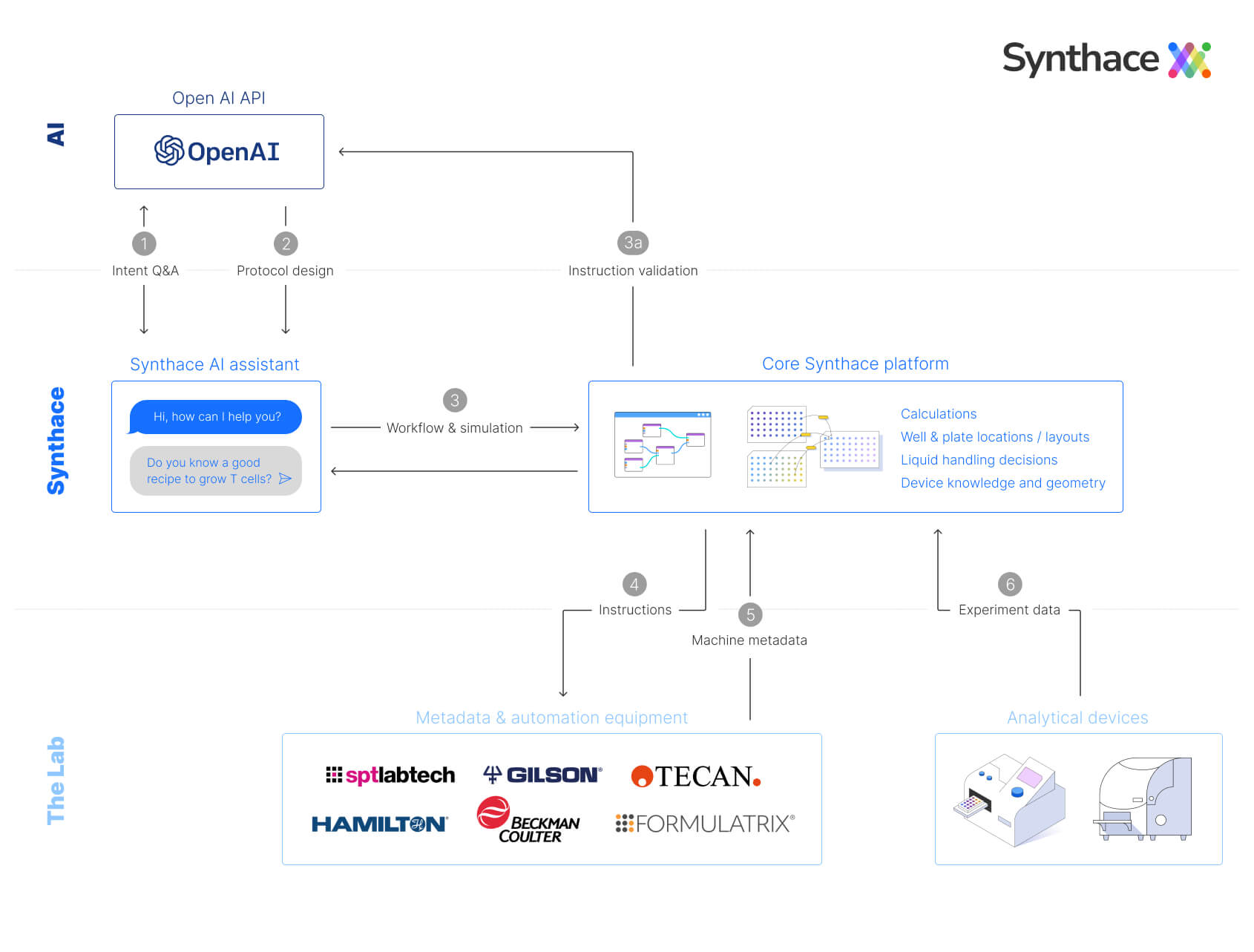
Schematic of the interface between OpenAI and Synthace
Between these APIs we have Python code that creates a context-aware prompt that precedes the user’s query with examples of a JSON data structure to be returned whenever the user asks for a protocol to be created. This format is a simplified, high-level description of what actions to perform.
This is then expanded into a full Synthace workflow and sent to the Synthace planner to simulate. If any errors occur it reports these back to ChatGPT to see if it can fix them. The example below shows an error message automatically and successfully fixed by ChatGPT by lowering the target volume without user input.
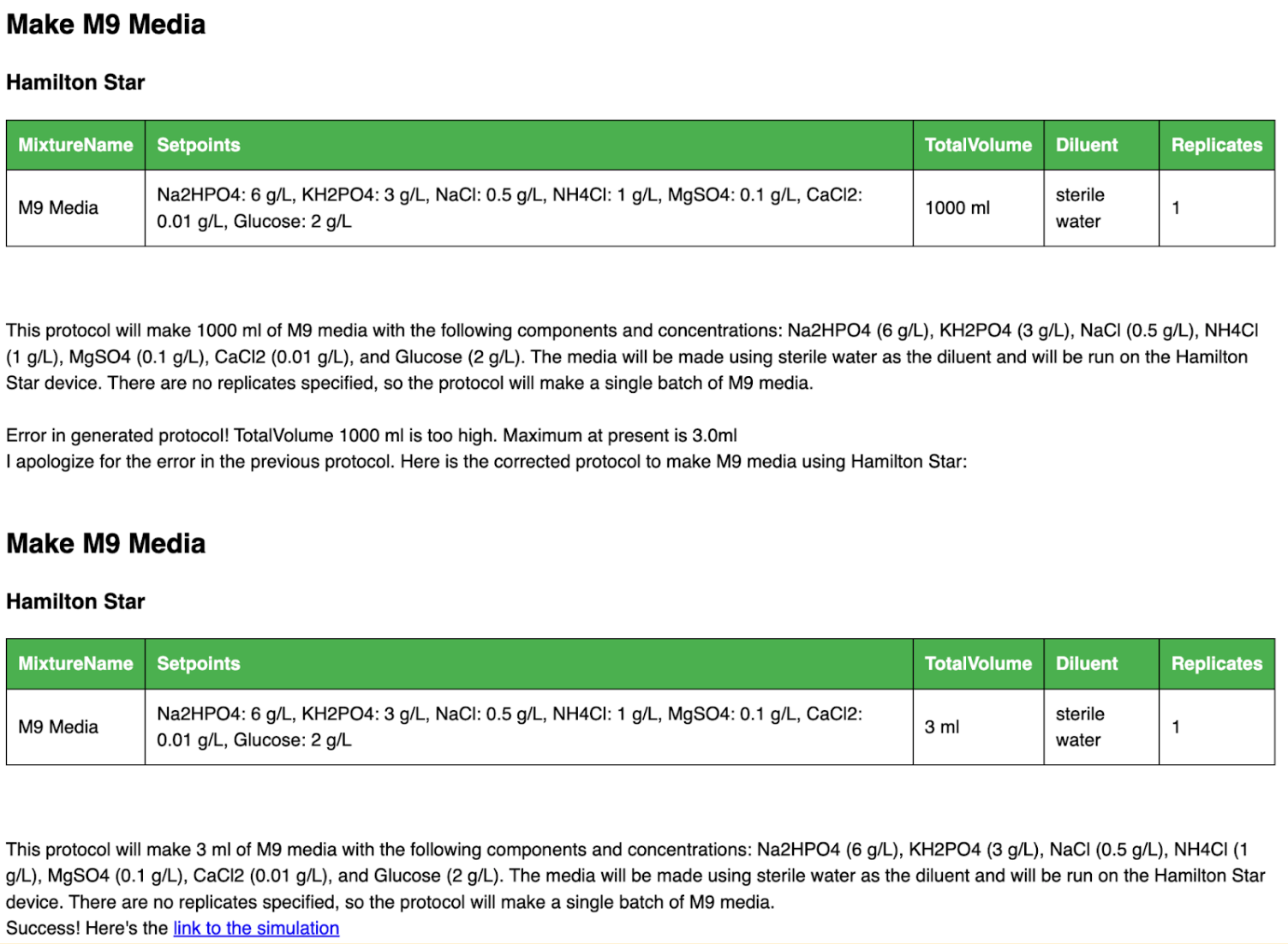
Example error showing total volume of reaction to be over 1000 mL and ChatGPT automatically correcting the method for a 3 mL total volume
How (and why) this integration works
As Stephen Wolfram mentions in his blog, “as an LLM neural net, ChatGPT—for all its remarkable prowess in textually generating material “like” what it’s read from the web, etc.—can’t itself be expected to do actual nontrivial computations, or to systematically produce correct (rather than just “looks roughly right”) data.”
The exact same case applies in our scenario. ChatGPT is brilliant at inferring intent from the user’s query; not just from a single interaction, but often from many inside the interface itself—a back-and-forth exploration. In many cases, it can return correct factual information, as we show in our example video (where a valid and commonly used growth media formulation is presented) which we use as the basis for our combinatorial media optimization experiment.
What ChatGPT is not good at is guaranteeing ground truth information. This is why Synthace checks the sanity of the answers it’s given because, sometimes, ChatGPT makes critical mistakes and contradictions. Instead of relying on it for everything, the power and greater robustness in this prototype comes from the delegation of appropriate planning tasks to the Synthace platform and the background interactions between the Python code, ChatGPT, and Synthace to avoid/fix errors. Despite this, mistakes in the generated protocols can still occur in which case there is additional opportunity to fix these with further user queries.
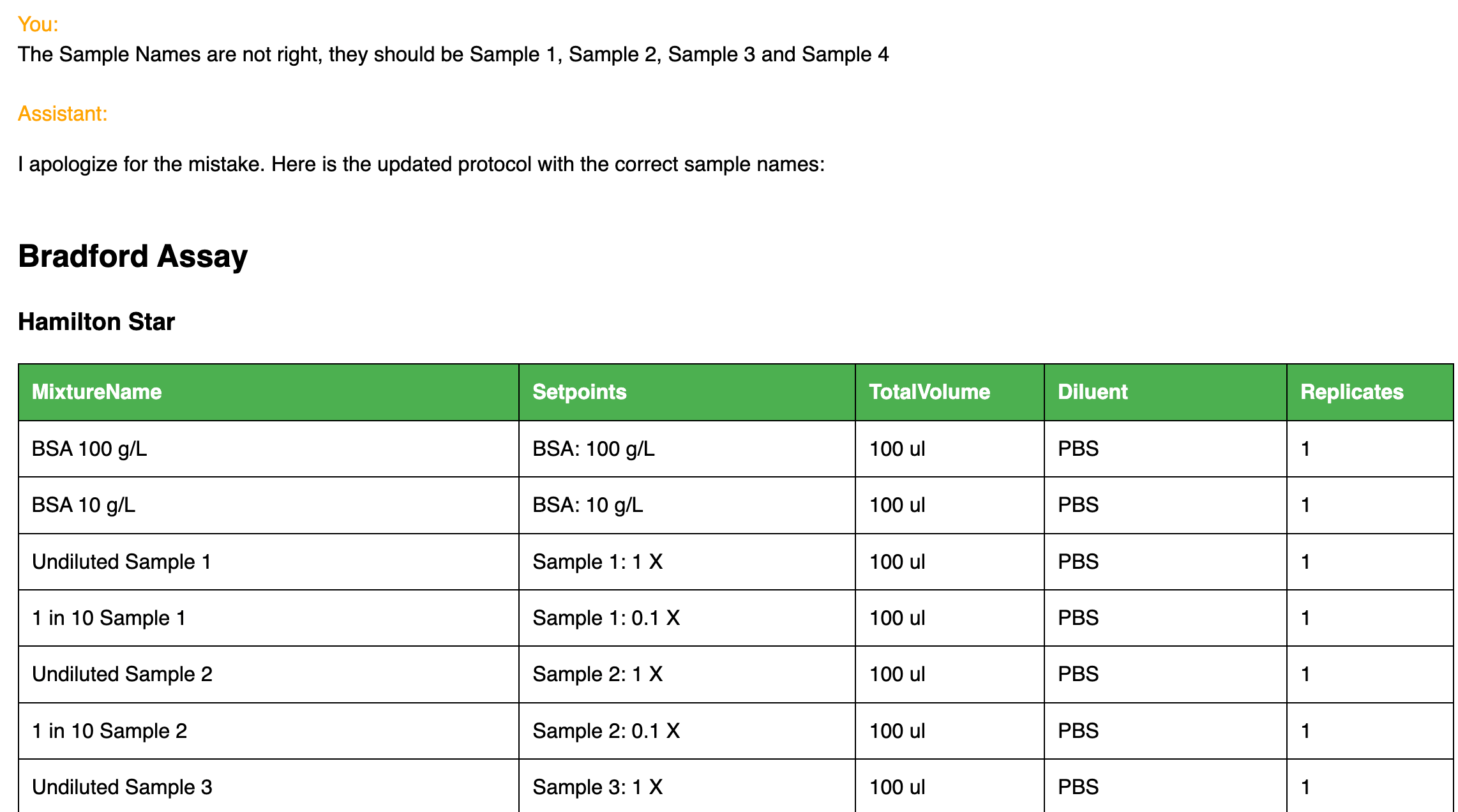
Making corrections with user queries
We try to avoid ChatGPT making calculations and doing math, and we avoid it supplying low-level execution details. In short: the more ChatGPT has to do, and the more complex the data structure it must return, the more there is to go wrong. Instead, we use Synthace to plan how best to execute each workflow and use ChatGPT to infer intent from natural language.
ChatGPT provides:
- Interpretation of the user’s experimental intent
- Guidance for the user to be able to specify any execution constraints and preferences such as incubations, addition order, and assays we want to perform
- A minimal schema describing the experimental requirements to send to Synthace
Synthace takes this intent and provides, in return:
- Calculations
- Well and plate location layouts
- Liquid handling decisions (volumes, stocks, and liquid class choices)
- Device knowledge and geometry
- Validation and feasibility of the experiment
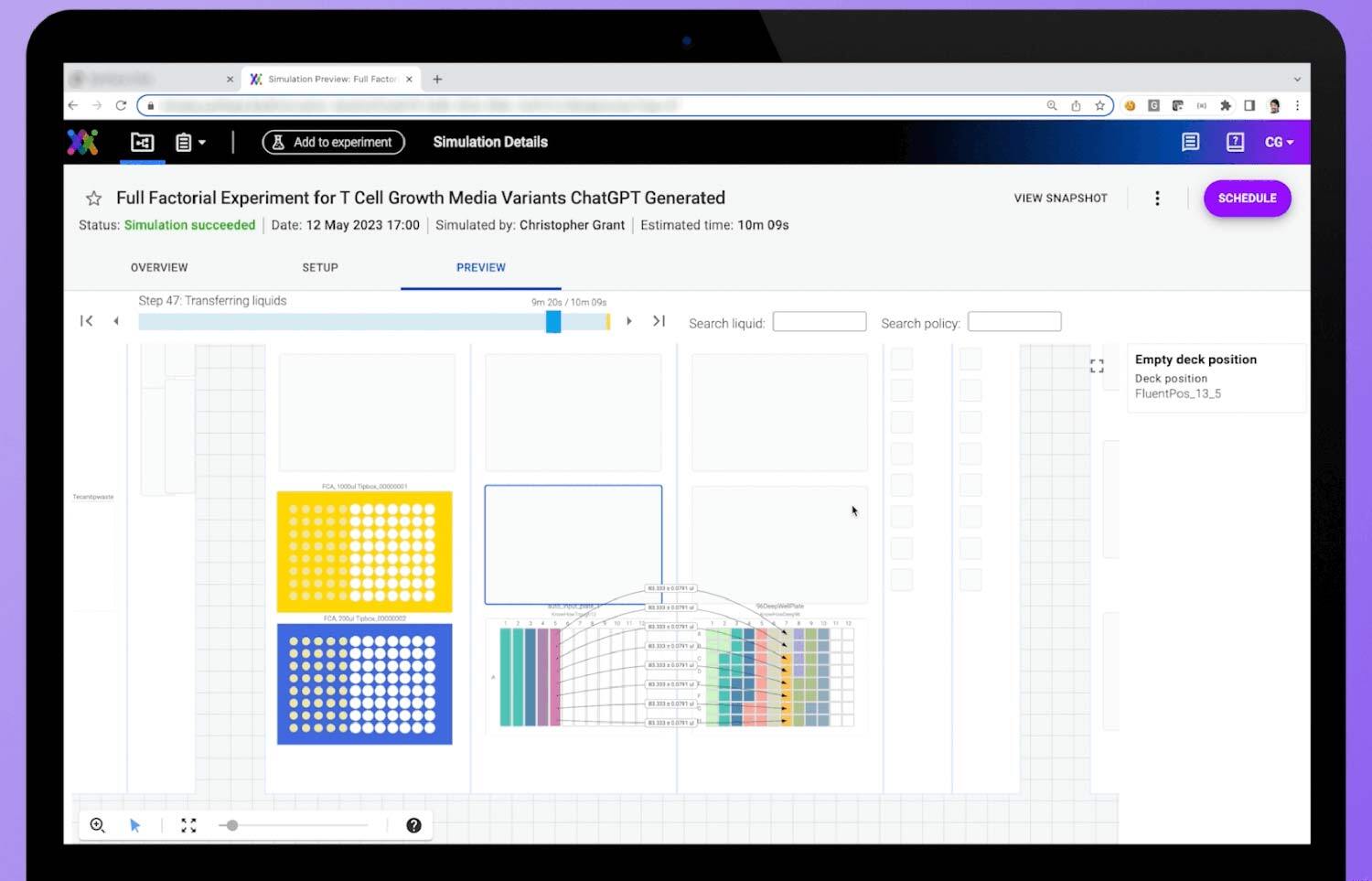
What a workflow simulation looks like on the Synthace platform
How does prompting work?
There are roughly three ways we can design and develop a protocol with the ChatGPT interface right now.
Give the AI all the necessary details in a single prompt:
Scientist: Create this Bradford Assay protocol in Synthace on a Tecan Fluent:
- Prepare a series of standard protein solutions with known concentrations of Bovine Serum Albumin (0.1, 0.3, and 0.5 mg/mL) by diluting BSA with PBS.
- Pipette 10 µL of each standard solution into a separate well of a microtitre plate in triplicate.
- Pipette 10 µL of an unknown sample into a separate well of the microtitre plate.
- Add 200 µL of Bradford reagent to each well, including the standards and the unknown sample.
Assistant: OK, here’s your protocol…
Step by step back-and-forth where the AI asks to for more information:
Scientist: Perform a Bradford assay in Synthace on a Hamilton Star
Assistant: I need some additional information please:
- What concentration range do you expect the protein to fall within?
- How many dilutions do you want to test between this range?
- What volumes of reagents should I use or should I look them up?
- What are the names of the protein samples you want to test?
Scientist:
- Between 0.001 ug/ul and 0.1 ug/ul
- 3
- Look them up
- Protein 1
Assistant: OK, here’s your protocol…
Let the AI make its own decisions:
Scientist: Perform a Bradford assay in Synthace on a Hamilton Star:
Assistant: I need some additional information please:
- What concentration range do you expect the protein to fall within?
- How many dilutions do you want to test between this range?
- What volumes of reagents should I use or should I look them up?
- What are the names of the protein samples you want to test?
Scientist: you decide
Assistant: OK, here’s your protocol…
An example below shows how “letting the AI decide” plays out. The AI assistant understands what a Bradford assay is (an assay to measure the protein content in a sample) and also knows what steps are needed, incubation and plate reader conditions, and that there are additional constraints compared to the media optimization example we show in the video. For example, in this case, the order of addition is important and so this is constrained.
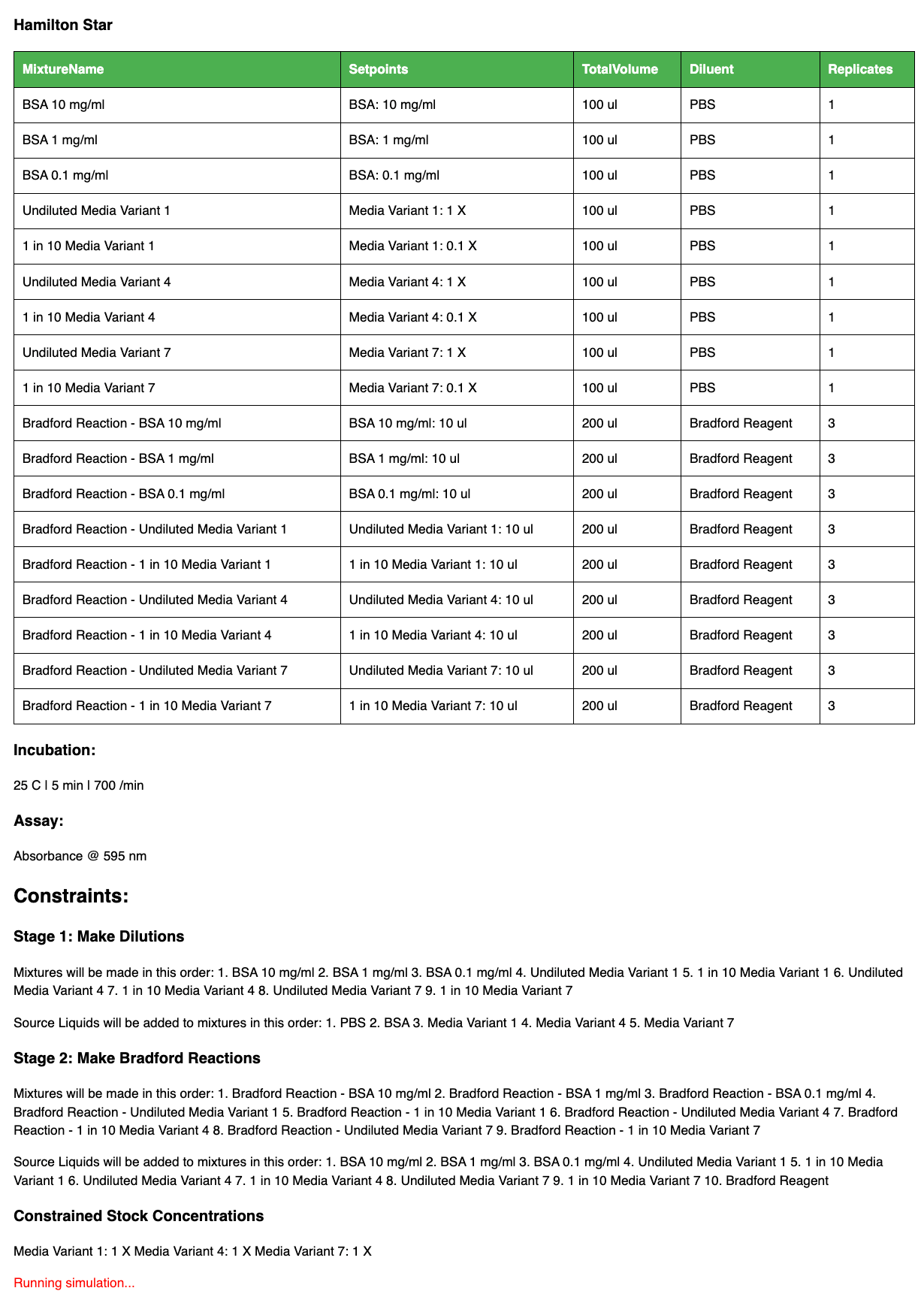
A full Bradford Assay created with ChatGPT and Synthace
What biological protocols are currently possible with ChatGPT?
The example shown in the video comes from a simple prompt for which media is suitable for growing T cells. There were sufficient examples in the training data upon which ChatGPT was trained for it to find a commonly used media recipe, with detailed composition and instructions on how to make it.
We also demonstrate that it knows what a Design of Experiments (DOE) full factorial design looks like and what we mean when we ask it to create one. We’ve also found it capable of knowing how to set up DNA processing protocols such as site-directed mutagenesis and CRISPR.
We’ve also found it can give reasonable instructions on how to set up common assays. For example, when asking for a protocol to quantify the protein in a sample, it returned a Bradford Assay protocol complete with controls and dilutions for creating a standard curve. While the returned results are not always perfect and should be double-checked by the user prior to running, it's impressive that this level of working protocol generation is achievable starting with a few vague prompts.
As for further/novel experiments, the AI assistant can infer meaning from the user's prompts to help understand what the user may want to do based on the GPT-3.5 training data, but is not restricted to just the protocols it has been trained on. This means, with some back-and-forth guidance by a scientist, it can generate experiments for new scenarios.
GPT-3.5 is, as far as we have been able to discern, trained on publicly available scientific method descriptions and papers. New models better than GPT-3.5 will likely be more powerful, especially models trained—in theory—on all scientific literature.
How we run the same protocol on multiple devices
Another key advantage that makes our platform particularly suited to integration with an AI assistant is that we can convert the same protocol to run on different automated devices. Liquid handling instructions are automatically optimized and can be executed for the equivalent protocol.
Right now, we can generate protocols for the following:
- Liquid handlers, e.g. from Tecan, Hamilton, or Gilson
- Liquid dispensers, e.g. the Beckman Coulter Echo, SPT Labtech Dragonfly, Gyger Certus Flex, or Formulatrix Mantis
- Manual pipetting instructions
The reason we can do this is because of our underlying digital experiment model and the work we’ve done integrating different automated liquid handling robots and dispensers into our platform over time.
Has anyone else done this already?
Using ChatGPT to run another program is no longer groundbreaking at this point, but as for this specific application, we’ve seen a variety of other developments at different levels of sophistication.
We’ve seen examples of ChatGPT writing machine code for the open-source OT2 liquid handler from Opentrons, as well as a coding assistant to program Hamilton liquid handlers, and as a kind of back-and-forth dialogue directing a scientist to set up an assay (again on an Opentrons), moving step by step. As far as we are aware, the use of Opentrons code in these examples relies on the code being open-source and available when GPT-3.5 was trained (up to 2021).
While ChatGPT is capable of generating the code shown in those examples, this approach suffers from a number of setbacks:
- Often wrong in subtle ways
- Device-specific
- Low-level (e.g. pick up tips, move liquid from A1 to B1 using this liquid class, etc)
- Limited by the size of the program that can be generated (because of the token limit)
- Requires a biologist to have a basic understanding of code
It’s also just naked code: it must be run, then the script uploaded to the device. Unlike our prototype, it’s not integrated into a cloud-based execution system. With Synthace, we can run the simulation remotely, connect to a device in a lab and push the protocol onto a device to be run.
A more advanced demonstration we have seen is in this paper, "Emergent autonomous scientific research capabilities of large language models.” The authors report on the development of a complex multi-component agent, based on LLMs developed to be capable of autonomous design, planning, and execution of scientific experiments from simple prompts. Particularly interesting aspects of their work included searching Google to get up-to-date information in order to design experiments, and querying device documentation and APIs in order to generate instructions for devices, even if the data was not included in the initial training data set for the LLM in question. They demonstrate this approach for generating Python code for running an Opentrons with a heater shaker module which was not available in the original GPT training set.
What this could mean in the near future
In their AI scientist grand challenge—a push to “develop AI systems capable of making Nobel quality scientific discoveries by 2050”—The Alan Turing Institute highlights “sensing and robotics” and “interfaces and interactive systems” as two of five key ways that AI might support scientists. While we don’t think we have a fully-fledged AI scientist just yet, this does seem to be a step in the right direction on both of these fronts.
Even with the current implementation of this interface, it offers an intuitive way for scientists to set up experimental protocols more easily, without them having to learn code or bespoke automation software. The barrier to using automation lowers drastically, the benefits of which have been well documented.
The barrier to running more powerful and insightful experiments using statistical methods, such as DOE, also lowers even further, with improved insight, reproducibility, and shareability. Ultimately, this means we can do better science and produce higher-quality data from which we can, in turn, learn much more.
This is just the starting point though. With models capable of handling larger volumes of text in both query and response, this opens the door for more complex protocol design and, possibly, uploading scientific papers to generate lab-executable protocols based on published literature.
Additionally, if a simulation is subsequently run in the lab, the Synthace platform and digital experiment model manages the results of that execution—all of the experiment data and metadata about how it was produced is captured. It’s conceivable that an AI assistant can also play a role in processing these results and generating reports from the experiment, perhaps then iterating further to run the next experiment and managing whole experimental campaigns.
There is also the possibility of a hybrid approach: using AI and carefully guided questioning in order to accurately and fully record and link experimental intent, planning, execution, results, interpretation, and distributing insights in a standardized, reproducible format that can be subsequently learned from and built upon in a way that is currently impossible.
All large language models are wrong, some are useful
As a company that was founded on the principles of DOE, I’m often reminded of the quote from George Box: “all models are wrong, some are useful.” Generative AI is also “just a model,” albeit with 175 billion parameters that predict the probabilities of the next words to follow in a sentence.
Both the impressive feats and hallucinations of ChatGPT have been widely reported showing that the George Box quote applies just as much to LLMs. Despite this, the approach looks extremely promising and, while it’s certainly no substitute for a human expert, large language models combined with expert scientists using our platform could generate and run experiments at a power, speed, and level of accuracy that we perhaps aren’t accustomed to yet.
Other frequently asked questions
- Is this part of the Synthace platform available today? No, this is only a prototype right now.
- Will you develop this further? Commercial interest is already prompting us to explore this further.
- Could anyone with ChatGPT and a liquid handler do the same thing? No. The only reason we can do this is because of the Synthace platform and its ability to represent, plan, validate, and automate accurate, flexible protocols.
- There's a lot of stories circling around the dangers of AI. Could this integration be harmful? Even in the event that we make this integration available on the Synthace platform, only users with a Synthace license would have access to this integration, which would enable us to minimize the risk of harmful use. Open AI, along with other makers of LLMs, are also investing in measures to counteract misuse and already have some safeguards in place.
- What could this mean for scientists? Scientists will likely be able to achieve much more in the lab, potentially significantly increasing the productivity of R&D in the life sciences.
- What would this mean for automation scientists and engineers?
Many have started to explore how ChatGPT can be useful in the lab automation tech stack. This showcases a baseline starting point for integration and can open up innovation towards other systems such as Electronic Lab notebooks (ELNs) and Laboratory Information Management Systems (LIMS). - What about the risk of misinformation and disinformation? It's no secret that LLMs are trained on data that hasn't been vetted or fact-checked (see Emily Bender and co-authors' paper on the topic), so it's important to be aware of potential misinformation, disinformation, and biases in the LLM's responses.
- What about sensitive data and intellectual property? While this integration is still only a prototype, taking an approach that protects data and IP will be vital for future commercial developments in this space. This is incredibly important to us and our customers. As for OpenAI, they state on their website that they do not “use data submitted by customers via our API to train OpenAI models or improve OpenAI’s service offering.”
Invitation to collaborate
While this is still a prototype, commercial interest is already prompting us to explore this further. We welcome involvement and collaboration from all groups. If you are interested in working together, please contact us with an outline of your interest and how you are interested in collaborating.
You can read our official press release here. We’ve uploaded the above video to YouTube, along with a post on LinkedIn, where you can follow us for more updates.
You can also subscribe to our newsletter at the bottom of this page to get notified of new posts.
About Synthace
Synthace is a digital experiment platform built for life science R&D teams to help them run more powerful experiments. It digitizes experiments from end to end: with minimal training, scientists can design and plan reproducible experiments, simulate them ahead of time, run them on their automation equipment, and automatically structure all of their experimental data and metadata in a single place. Controlled from a browser window and needing no code to operate, it makes experiments easier to run in the lab, increases the scientific value of experiments, and also makes it possible to run experiments that were previously thought impossible.
Already deployed in 9 of the top 20 life science companies around the world, the Synthace platform fundamentally transforms and improves the relationship between what a scientist can imagine and what they can actually do in their lab.
Media contact
Please contact Will Patrick, Synthace Director of Brand and Communications: will.patrick@synthace.com
Chris Grant, PhD
Dr Chris Grant is the Head of Research and Co-founder of Synthace. Chris has been at the forefront of Synthace’s development, applying his knowledge in bioprocessing, synthetic biology, lab automation, software engineering, and machine learning to help shape the future of the product.


